



University of Rostock
Department of Anatomy
Neuroscience Group


neuroVIISAS - Connectivity matrices

←
→
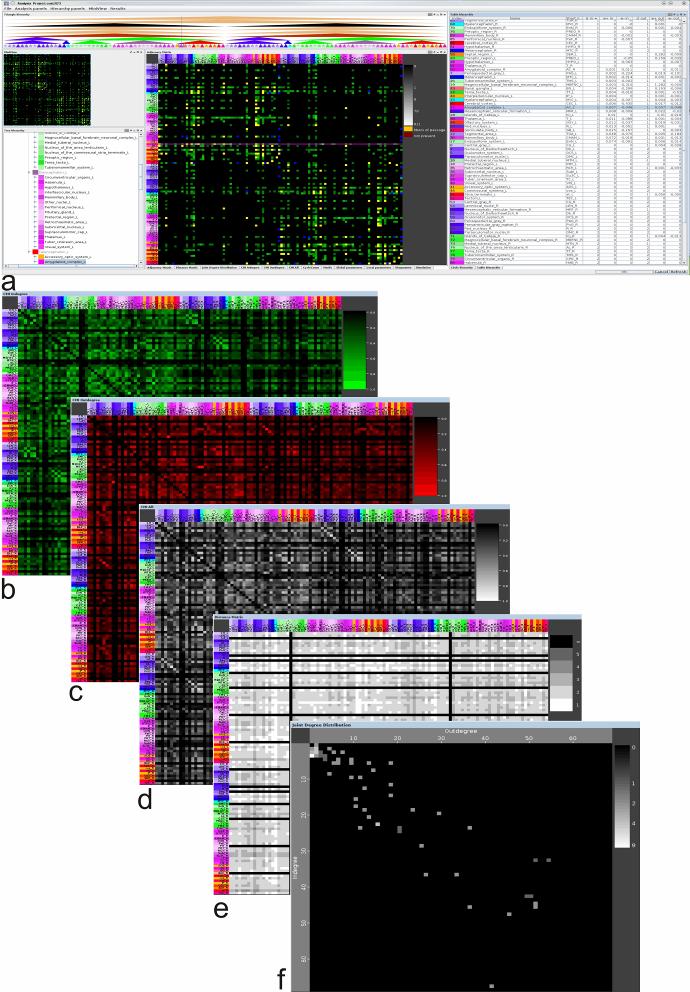
Graph theory based analysis of connectivities. The following matrices
(figure with overview of matrices is shown below) and parameters are
computed:
- Weighted and non-weighted adjacency matrices
- Clustermatching index matrix for indegrees
- Clustermatching index matrix for outdegrees
- Clustermatching index matrix for in- and outdegrees
- Distance matrix
- Joint degree distribution.
Furthermore, the generalized topology overlaping measure matrix
(GTOM), spatial distance matrix and communicability matrix can be
computed, cycle count lists can be generated and statistical motif /
subgraph analysis can be performed.

Homepage |
Staff |
neuroVIISAS |
Rat connectome |
Other connectomes |
Neuroimaging |
Neurodegeneration |
Students |
Links |
Publications






