



University of Rostock
Department of Anatomy
Neuroscience Group


neuroVIISAS - Local network parameters
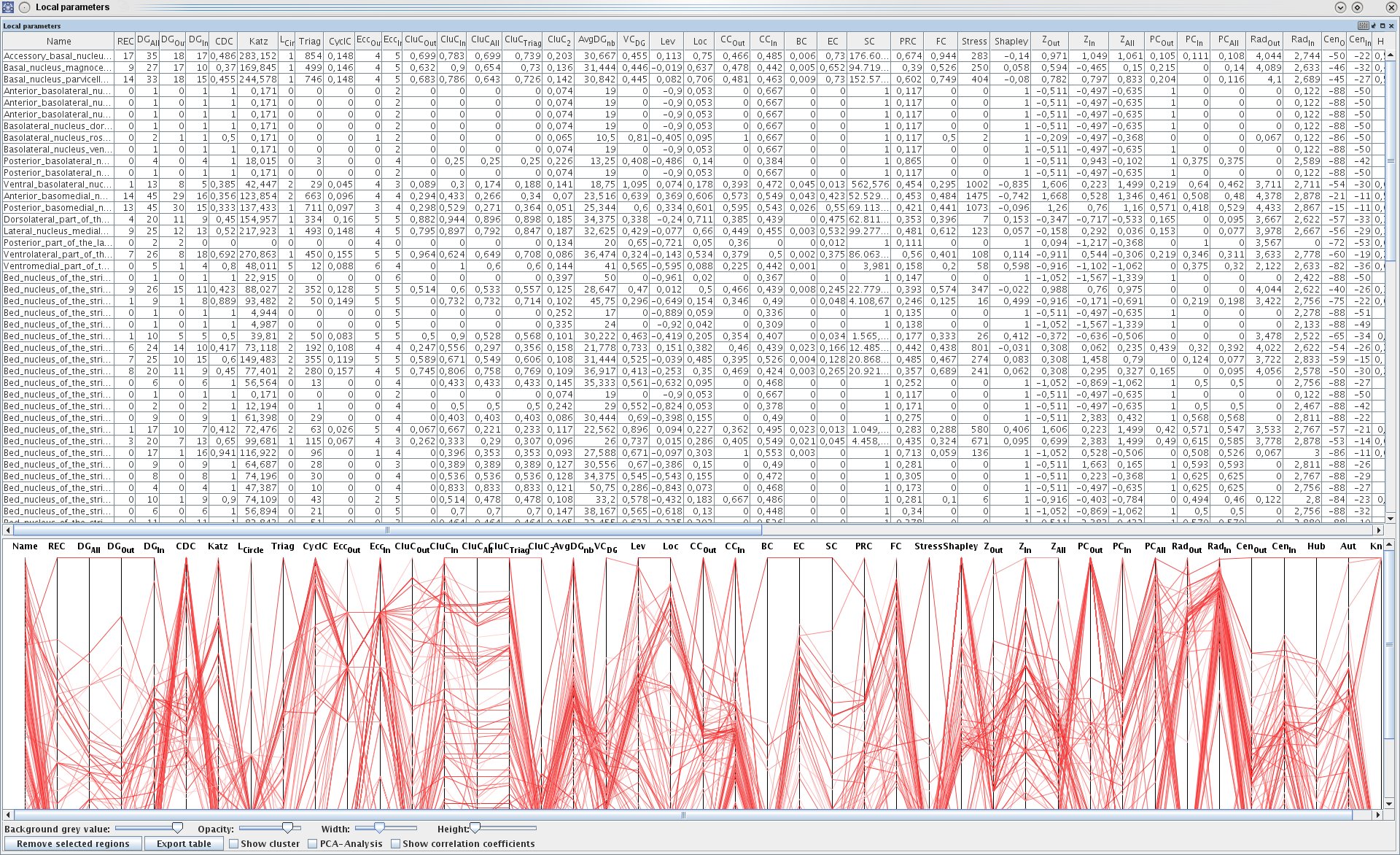
Local network parameters and a parallel coordinate visualization (visual analytics). HD: hierarchy depth, REC: umber of reciprocal edges, DG_all: Indegree + Outdegree, DG_out: Outdegree, DG_in: Indegree, CDC: Convergence-Divergence_Coefficient, Lat: Laterality, Katz: Katz status index, L_Circle: Shortest path to a node, Triag: Number of triangles around a node, CyclC: Cyclic coefficient, ECC_Out: Excentricity out, ECC_In: Excentricity in, CCoeff_out: Clustercoefficient out, CCoeff_in: Clustercoefficient in, CCoeff_all: Clustercoefficient in and out, CluC_Triag: Clustercoefficient triangle based, CluC_2: Hierarchical clustercoefficient of level 2 (second, indirect neighbours), AvgDG_nb: Arithmetic mean of the degree all of all neighbors of a node, VC_DG: Variation coefficient of the neighbor degrees, Lev: Leverage, Loc: Locality index, CC_out: Closeness centrality out, CC_in: Closeness centrality in, BC: Betweeness centrality, EC: Eigenvector centrality, SC: Subgraph centrality, PRC: Page rang centrality, FC: Flow coefficient, Stress: Number of shortest paths through a node, Shapley rating, Z_out/in/all: Z-score, PC_out/in/all: Participation coefficient, Rad_out/in: Radiality, Cen_out/in: Centroid value, Hub: Hubness, Aut: Authoritativeness, Knot: Knotty centrality